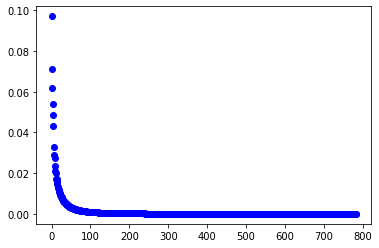
PCA의 효과를 알아보기 위해 약 95%의 variance를 차지하는 principal component들을 골라 MNIST dataset에 대해 비교해보았습니다. 공백이 많은 image data인 만큼 784개의 feature(pixel)들이 고르게 분산(explained_variable_ratio_)을 가지고 있지 않았습니다. PCA를 적용하기 적절한 data죠.
원래의 784개의 feature들과 PCA를 적용하여 선택된 상위 몇 개의 principal component들을 동일한 모델의 input으로 사용하여 결과를 비교했습니다. (2개의 모델을 사용하여 실험)
그 결과, PCA를 적용한 data의 경우, 쓸데없는 정보를 가진 feature의 개수가 더 적어 model의 분산이 낮아져 overfitting을 방지하는 효과를 보였습니다.
import tensorflow as tf
import numpy as np
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Dropout, BatchNormalization, Activation
import matplotlib.pyplot as plt
%matplotlib inline
mnist = tf.keras.datasets.mnist
(X_train, y_train), (X_test, y_test) = mnist.load_data()
X_train, X_test = X_train.reshape(-1, 784), X_test.reshape(-1, 784)
pca = PCA(86)
X_train_pca = pca.fit_transform(X_train)
X_test_pca = pca.transform(X_test)
std_scaler = StandardScaler()
X_train_pca = std_scaler.fit_transform(X_train_pca)
X_test_pca = std_scaler.transform(X_test_pca)
X_train = std_scaler.fit_transform(X_train)
X_test = std_scaler.transform(X_test)
var = pca.explained_variance_ratio_
plt.plot(var, 'bo');
cum_var = 0
for i, v in enumerate(var):
cum_var = cum_var + v
# if cum_var > 0.90:
# print("90% variance:", i) # 86
if cum_var > 0.95:
print("95% variance:", i) # 153
break
95% variance: 153
def generate_model(input_dim):
model = Sequential([
Dense(512, kernel_initializer='he_normal', input_dim=input_dim),
BatchNormalization(), Activation('elu'), Dropout(0.1),
Dense(512, kernel_initializer='he_normal', input_dim=784),
BatchNormalization(), Activation('elu'), Dropout(0.1),
Dense(10, activation='softmax')
])
return model
model = generate_model(X_train.shape[-1])
model.compile(loss='sparse_categorical_crossentropy', optimizer='adam', metrics=['acc'])
h = model.fit(X_train, y_train, epochs=100, validation_split=0.2, verbose=0)
model_pca = generate_model(X_train_pca.shape[-1])
model_pca.compile(loss='sparse_categorical_crossentropy', optimizer='adam', metrics=['acc'])
h_pca = model_pca.fit(X_train_pca, y_train, epochs=100, validation_split=0.2, verbose=0)
train_loss, val_loss = h.history['loss'], h.history['val_loss']
train_pca_loss, val_pca_loss = h_pca.history['loss'], h_pca.history['val_loss']
plt.figure(figsize=(15, 5))
plt.plot(train_loss, 'b-', label='train'); plt.plot(val_loss, 'b--', label='val')
plt.plot(train_pca_loss, 'r-', label='train_pca'); plt.plot(val_pca_loss, 'r--', label='val_pca')
plt.title('PCA comparation'); plt.xlabel('epochs'); plt.ylabel('loss'); plt.ylim([0, 0.35]);
plt.grid(); plt.legend();
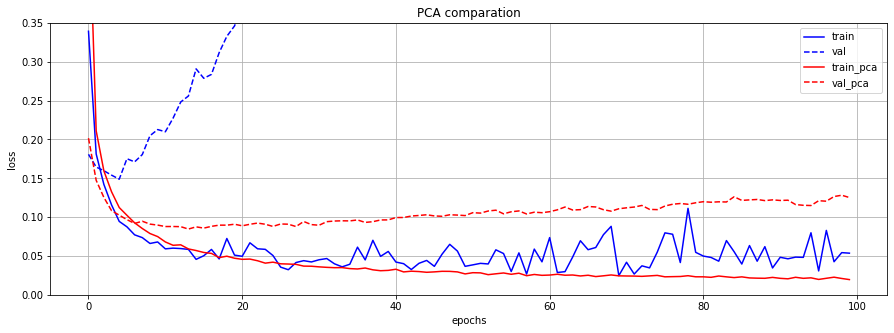
Tensorflow2 Tutorial Model1
def generate_model(input_dim):
model = Sequential([
Dense(128, activation='relu', input_dim=input_dim),
Dropout(0.2),
Dense(10, activation='softmax')
])
return model
model = generate_model(X_train.shape[-1])
model.compile(loss='sparse_categorical_crossentropy', optimizer='adam', metrics=['acc'])
h = model.fit(X_train, y_train, epochs=100, validation_split=0.2, verbose=0)
model_pca = generate_model(X_train_pca.shape[-1])
model_pca.compile(loss='sparse_categorical_crossentropy', optimizer='adam', metrics=['acc'])
h_pca = model_pca.fit(X_train_pca, y_train, epochs=100, validation_split=0.2, verbose=0)
train_loss, val_loss = h.history['loss'], h.history['val_loss']
train_pca_loss, val_pca_loss = h_pca.history['loss'], h_pca.history['val_loss']
plt.figure(figsize=(15, 5))
plt.plot(train_loss, 'b-', label='train'); plt.plot(val_loss, 'b--', label='val')
plt.plot(train_pca_loss, 'r-', label='train_pca'); plt.plot(val_pca_loss, 'r--', label='val_pca')
plt.title('PCA comparation'); plt.xlabel('epochs'); plt.ylabel('loss'); plt.ylim([0, 0.35]);
plt.grid(); plt.legend();
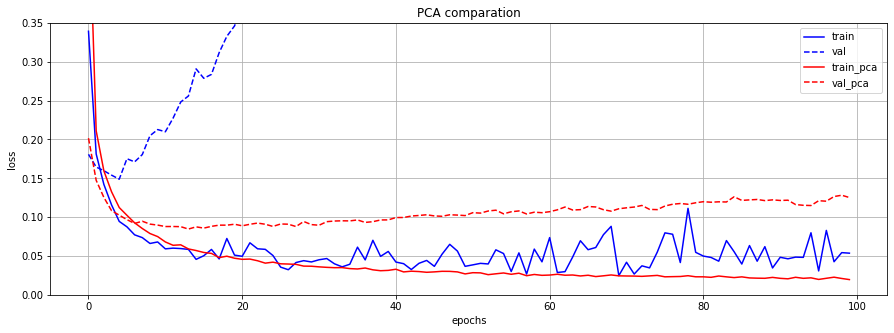
PREVIOUSEtc